Evaluation of the RESTful API for molar to mass concentration conversion using UCUM and LOINC¶
Sam Tomioka
Last Update May 26, 2019
TOC:¶
- Introduction
- Findings
- 2-1. Prior Work
- 2-2. Findings on Updated UCUM Conversion (May 5, 2019)
- 2-2-1. Conversion Results
- 2-2-2. Error Messages from UCUM Conversion
- 2-2-3. Summary
- 2-3. Findings on updated UCUM service (May 26, 2019)
- 2-3-1 Conversion Results
- 2-3-2 Error Messages from UCUM Conversion
- 2-3-3 Summary
- Scripts
- Output Issues
- 4-1. Discrepancies between LBSTRESN and Conversion based on UCUM
- 4-2. Check differences between reported LBSTRESN vs Converted Results
- 4-2-1. Normalized mean differences between souce and UCUM
- 4-2-2. Normalized maximum differences between souce and UCUM
- 4-2-3. Normalized minimum differences between souce and UCUM
- 4-2-4. Compare mean results vs. mean differences
- 4-2-5. Check distributions of differences between the source and UCUM conversion
- 4-2-6. Summary statistics of the source and UCUM conversion on $\text{Mg}^{+2}$
- 4-2-7. Summary statistics of the source and UCUM conversion on PHOS
- 4-2-8. Summary statistics of the source and UCUM conversion on TSH
- 4-2-9. Summary statistics of the source and UCUM conversion on BILI
- 4-2-10. Summary statistics of the source and UCUM conversion on MONOLE
- 4-2-11. Summary statistics of the source and UCUM conversion on HBA1C
- 4-3-1. Summary of Error Message finding
1. Intruduction ¶
The verification of scientific units and conversion from the reported units to standard units have been always challenging for Data Science due to several reasons:
- Need a lookup table that consists of all possible input and output units for measurements, name of the measurements (e.g. Glucose, Weight, ...), conversion factors, molar weights etc.
- The names of the measurement in the lookup table and incoming data must match
- The incoming units must be in the lookup table
- Maintenance of the lookup table must be synched with standard terminology update
- Require careful medical review in addition to laborsome Data Science review and more...
Despite the challenges, the lookup table approach is the norm for many companies for verification of the units and conversion. Consideration was given for more systematic approach that does not require to use the lab test names[1], but some units rely on molar weight and/or valence of ion of the specific lab tests, so this approach does not solve the problem. The regulatory agencies require the sponsor to use standardized units for reporting and analysis[2]. The PMDA requires SI units for all reporting and analysis[3,4]. The differences in requirement force us to maintain region specific conversion for some measurements which add additional complexity.
The approach Jozef Aerts discussed uses RestAPI available through Unified Code for Units of Measure (UCUM) Resources which is maintained by the US National Library of Medicine (NLM)[5]. The benefit is obvious that we can potentially eliminate the maintenance of the lab conversion lookup table. Here is what they say about themselves.
The Unified Code for Units of Measure (UCUM) is a code system intended to include all units of measures being contemporarily used in international science, engineering, and business. The purpose is to facilitate unambiguous electronic communication of quantities together with their units. The focus is on electronic communication, as opposed to communication between humans. A typical application of The Unified Code for Units of Measure are electronic data interchange (EDI) protocols, but there is nothing that prevents it from being used in other types of machine communication.
The UCUM is the ISO 11240 compliant standard and has been used in ICSR E2B submissions for regulators adopted ICH E2B(R3). FDA requires the UCUM codes for the eVAERS ICSR E2B (R3) submissions, dosage strength in both content of product labeling and Drug Establishment Registration and Drug Listing. UCUM codes have been adopted by HL7 FHIR.
1-1. Introduction for mol-mass/mass-mol conversion ¶
Jozef Aerts announced an updated RESTful API which accounts for the molecular weights of the analyte into the conversion between molar and mass concentrations. This additional functionality would facilitate the conversion of the lab results, verification of the standardized lab results and LOINC code provided by the vendors.
Although CDISC released a downloadable CDISC UNIT and UCUM mapping xlsx file, this evaluation will not use it since the CDISC UNIT does not cover all reported units used by the clinical laboratory/bioanalytical/PK vendors. Regular expression along with UCUM unit validity service was used to convert and verify the units provided by the lab vendors. In the future, this will be done with encoder-decoder or transformer + sequence-to-sequence model which demonstrated near perfect to generate iso 8601 from numerous date formats.
An initial evaluation was done on RestAPI available through the Unified Code for Units of Measure (UCUM) Resources and the findings are summarized in 2-1. The second evaluation is completed on the test version of RestAPI provided by Jozef Aerts at xml4pharma
2. Findings ¶
2-1. Prior Work ¶
Previoiusly the production version of RestAPI provided by US National Library of Medicine was evaluated. See here for more detail.
6458 laboratory records were used to test UCUM RestAPI. These records are from one of the ongoing clinical trial with standard set of clinical laboratory tests. Out of 6458 records, there were 321 records identified as incorrect conversions. Out of 322 findings, 169 was false positive which is due to lack of accounting valence of ion with respect to mEq to molar unit conversion.
| Records | |
|---|---|
| Total Records | 6458 |
| Identified as incorrect conversion | 321 |
| True Positive | 153 |
| False Positive | 169 |
2142 records were identified as error. Out of 2142 errors, 120 records identified as error due to having a categorical data despite unit was given. There were 2022 records where the source and target unit do not have the same property. Most of them are cause by lack of mass-mol conversions, and the rest appeared to be correct but medical judgement would be neccessary.
| Type of Error | Records |
|---|---|
| ERROR: unexpected result: Error: Source and Target unit do not seem to belong to the same property | 2022 |
| ERROR: unexpected result: NEGATIVE is not a numeric value | 119 |
| ERROR: unexpected result: Negative is not a numeric value | 1 |
Overall, this approach worked for majority of the records 6458, however, a few improvements are required by NLM/NIH to full utilize this RestAPI.
- Need for mass-mol conversions *
- Need to account for valence of ion with respect to mEq to molar unit conversion
- Need for an option to specify molar weight or LOINC code for accurate unit conversion with respect to molar unit.
2-2. Findings on Updated UCUM Conversion (May 5, 2019) ¶
A total of 419103 laboratory records were obtained from 17 clinical trials. Following steps were taken to reduce the number of records for the evaluation of a test version of UCUM Conversion API.
- Removal of records with missing units before and after UCUM unit conversion
- Removal of character type results such as ['Negative','None','Trace',...]
- Removal of records does not require conversion. For example, the records with LBORRESU==LBSTRSU were removed.
- Removal of duplicate records
17384 records of laboratory results were used for the evaluation. This evaluation does not cover the use of MOLWEIGHT.
The Table 3 below summarizes the number of records from each step.
| - | Number of Records |
|---|---|
| Number of studies | 17 |
| Input data | 419103 |
| After removal of missing units before UCUM unit conversion | 276144 |
| After removal of character results | 255937 |
| After removal of records do not require conversion | 172205 |
| After removal of duplicate records* | 17550 |
| After removal of missing units after UCUM unit conversion | 17384 |
2-2-1. Conversion Results ¶
There were 2620 records from 27 tests where the LBSTRESN and UCUM conversion results did not match. Observed differences are plotted in Section 4-1.
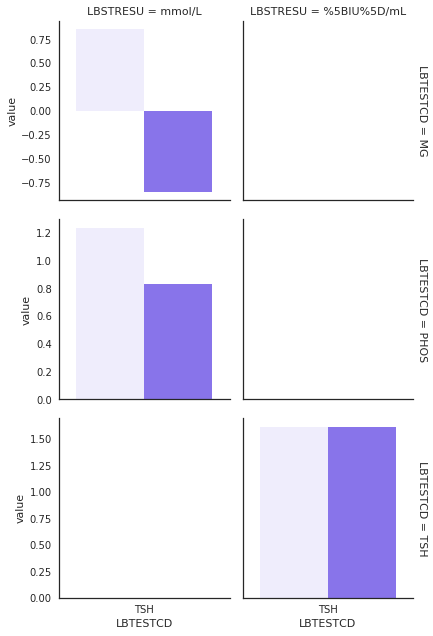
Out of 27 tests,PHOS, TSH, and MG had a large difference between the LBSTRESN and UCUM conversion. PHOS (n=72) and TSH (n=139) had true positive findings. One test, MG (n=20), had false positive findings. In Figure 1, the left light colored bars show the LBSTRESN, and the right dark colored bars show the difference between LBSTRESN and the returned value from UCUM conversion. The details are discussed in Sec 4-1, but the Table 4 summarizes the findings on these 3 tests.
| LBTESTCD | Source of Issue | My Note |
|---|---|---|
| MG | UCUM API | ion channel is ignored in conversion |
| PHOS | Input Data | mass-molar conversion was done incorrectly by the lab vendor |
| TSH | Input Data | mass-molar conversion was done incorrectly by the lab vendor |
2-2-2 Error Messages from UCUM Conversion ¶
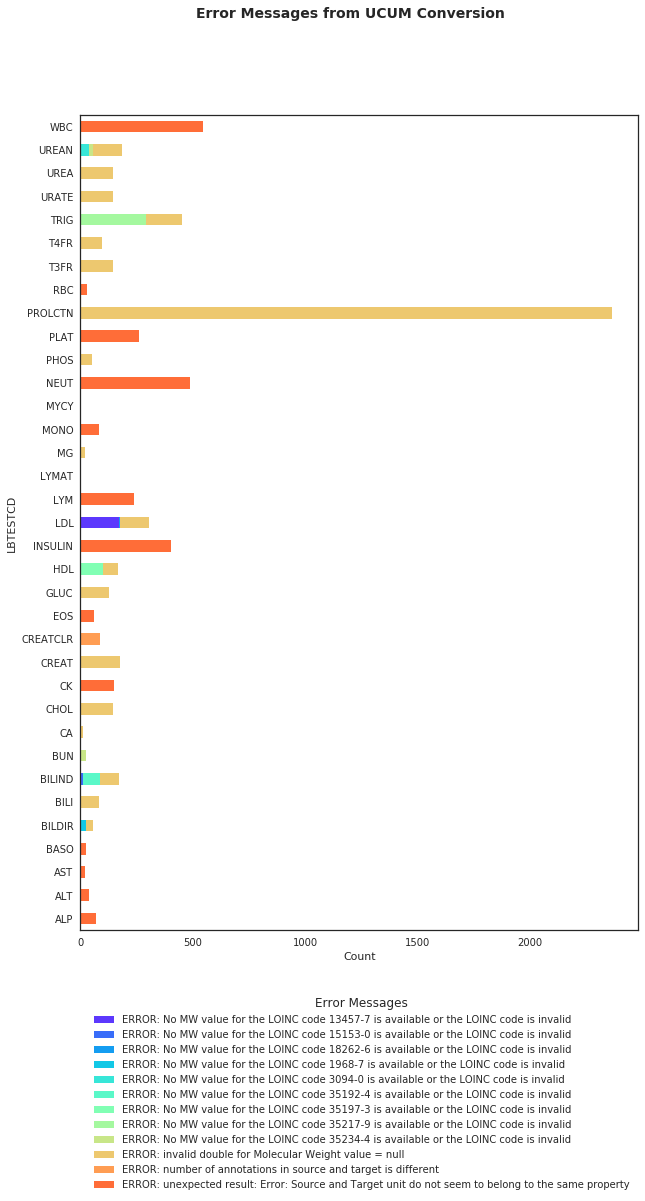
7389 records were returned with error messages from UCUM Conversion as shown in the Figure 2. Table 5 summarizes the type of errors received.
- The most frequent error was
ERROR: invalid double for Molecular Weight value = nullwhich turns out the be true positive finding. This is related to missing m.w. or LOINC when the conversion requires m.w.. - There were 9 kinds of
ERROR: No MW value for the LOINC code **xxxxxxx** is available or the LOINC code is invalid. One of the error was due to invalid LOINC code15153-0, but the rest of errors appear to be due to missing m.w. in the LOINC database. It would be helpful if this error message is split into each condition (1. invalid LOINC code or missing MW in LOINC) for our verification purpose. - Creatinine Clearance (mL/min/1.73 m2) conversion failed with
ERROR: number of annotations in source and target is different. The surface area 1.73 m2 was added as annotation for the source but the target did not include the same annotation. This will be solve with http://xml4pharmaserver.com:8080/UCUMService2/rest/ucumtransform/121/from/mL/min/%7B1.73_m2%7D/to/mL/s/%7B1.73_m2%7D - Several conversions failed with
ERROR: unexpected result: Error: Source and Target unit do not seem to belong to the same property. See Table 6 for more detail.
| Message | My Note | Sample Call |
|---|---|---|
| ERROR: invalid double for Molecular Weight value = null | True positive finding | http://xml4pharmaserver.com:8080/UCUMService2/rest/ucumtransform/0.3/from/mg/dL/to/umol/L |
| ERROR: No MW value for the LOINC code 13457-7 is available or the LOINC code is invalid | Valid LOINC, No m.w from LOINC | http://xml4pharmaserver.com:8080/UCUMService2/rest/ucumtransform/103/from/mg/dL/to/mmol/L/LOINC/13457-7 |
| ERROR: No MW value for the LOINC code 15153-0 is available or the LOINC code is invalid | Invalid LOINC | http://xml4pharmaserver.com:8080/UCUMService2/rest/ucumtransform/0.3/from/mg/dL/to/umol/L/LOINC/15153-0 |
| ERROR: No MW value for the LOINC code 18262-6 is available or the LOINC code is invalid | Valid LOINC, No m.w from LOINC | http://xml4pharmaserver.com:8080/UCUMService2/rest/ucumtransform/118/from/mg/dL/to/mmol/L/LOINC/18262-6 |
| ERROR: No MW value for the LOINC code 1968-7 is available or the LOINC code is invalid | Valid LOINC, No m.w from LOINC | http://xml4pharmaserver.com:8080/UCUMService2/rest/ucumtransform/0.1/from/mg/dL/to/umol/L/LOINC/1968-7 |
| ERROR: No MW value for the LOINC code 3094-0 is available or the LOINC code is invalid | Valid LOINC, No m.w from LOINC | http://xml4pharmaserver.com:8080/UCUMService2/rest/ucumtransform/10/from/mg/dL/to/mmol/L/LOINC/3094-0 |
| ERROR: No MW value for the LOINC code 35192-4 is available or the LOINC code is invalid | Valid LOINC, No m.w from LOINC | http://xml4pharmaserver.com:8080/UCUMService2/rest/ucumtransform/1.07/from/mg/dL/to/umol/L/LOINC/35192-4 |
| ERROR: No MW value for the LOINC code 35197-3 is available or the LOINC code is invalid | Valid LOINC, No m.w from LOINC | http://xml4pharmaserver.com:8080/UCUMService2/rest/ucumtransform/85/from/mg/dL/to/mmol/L/LOINC/35197-3 |
| ERROR: No MW value for the LOINC code 35217-9 is available or the LOINC code is invalid | Valid LOINC, No m.w from LOINC | http://xml4pharmaserver.com:8080/UCUMService2/rest/ucumtransform/42/from/mg/dL/to/mmol/L/LOINC/35217-9 |
| ERROR: No MW value for the LOINC code 35234-4 is available or the LOINC code is invalid | Valid LOINC, No m.w from LOINC | http://xml4pharmaserver.com:8080/UCUMService2/rest/ucumtransform/12/from/mg/dL/to/mmol/L/LOINC/35234-4 |
| ERROR: number of annotations in source and target is different | ?? | http://xml4pharmaserver.com:8080/UCUMService2/rest/ucumtransform/121/from/mL/min/%7B1.73_m2%7D/to/mL/s |
| ERROR: unexpected result: Error: Source and Target unit do not seem to belong to the same property | Why the error is not 'ERROR: invalid double for Molecular Weight value = null' | http://xml4pharmaserver.com:8080/UCUMService2/rest/ucumtransform/12.9/from/uU/mL/to/pmol/L |
The following converions as listed in the Table 6 returend the following error message.
ERROR: unexpected result: Error: Source and Target unit do not seem to belong to the same property
| LBTESTCD | LBORRESU | LBSTRESU |
|---|---|---|
| INSULIN | uU/mL | pmol/L |
| BASO | G/L | 10*9/L |
| EOS | G/L | 10*9/L |
| LYM | G/L | 10*9/L |
| MONO | G/L | 10*9/L |
| NEUT | G/L | 10*9/L |
| PLAT | G/L | 10*9/L |
| RBC | T/L | 10*12/L |
| WBC | G/L | 10*9/L |
| LYMAT | G/L | 10*9/L |
| MYCY | G/L | 10*9/L |
| ALP | %5BIU%5D/L | U/L |
| ALT | %5BIU%5D/L | U/L |
| AST | %5BIU%5D/L | U/L |
| CK | %5BIU%5D/L | U/L |
| INSULIN | u%5BIU%5D/mL | pmol/L |
GorGiga per literis often used in the hematology panel and is equivalent to 10*9 as per ucum-essence.xml, but the conversion was not sucessful.IUandUare equivalent, but the conversion failed.Tand `10*12' are equivalent as per ucum-essence.xml, but the conversion failed.
G |
U and IU |
T |
|---|---|---|
 |
 |
 |
Source:ucum-essence.xml
2-2-3. Summary ¶
This approach has potential and can be used for any units (PK, Lab, ECG, Vital Signs, etc). The addition of mass-mol/mol-mass conversion is a great addition and very useful to verify the results obtained from the vendors.
A few improvements would allow us to use this API at full potential.
As previously discusses, the varence of an ion with respect to mEq to molar unit conversion was one of the issues. Some LOINC based conversion was not performed due to lack of m.w.. Conversion for units with G, T,IU, and U were not successful.
'ERROR: No MW value for the LOINC code xxxxxxx is available or the LOINC code is invalid' could be split into two errors for each condition for verification purpose. Otherwise, one need to lookup LOINC to confirm whether or not the LOINC is valid or m.w. is missing.
Overall, the tool was very useful and we found the conversion issues caused by two vendors affecting many clinical trials. We will implement this in SAS for programmers, and Python for automated checking using the production release by NIH.
2-3. Findings on updated UCUM service (May 26, 2019) ¶
Since the previous review, additional updates were made to the UCUM service.
- The return message contains the MW that was used for the conversion.

The error message related to LOINC was split into two different message to reflect the actual issue. Previously, the error message was given as
Error message "ERROR: No MW value for the LOINC code xxx-x is available or the LOINC code is invalid"
The updated service returns LOINC related message either
- Invalid LOINC code XXXX
- No MW found for LOINC Part Number LPxxxx for LOINC code yyyy This updates allow us to investigate issues without browsing LOINC.
- The list of MW for the LOINC-component-part was extended
- The conversion that involves 'Eq' to molar unit now account for valence of ion when LOINC is provided. This should eliminate the conversion issues found previously for magnesium.
2-3-1. Conversion Results ¶
17565 lab records were tested for this review. All are unique records based on LBTESTCD, LBORRES, LBORRESU, LBSTRESU.
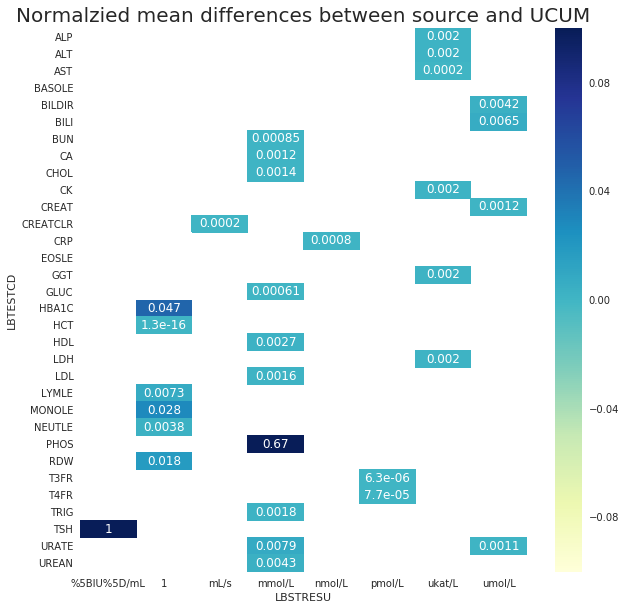
The following Figure 3 shows the normalized differences between the source and the UCUM conversions when they are not equal. There are many tests with negligible differences as I saw in the previous test. HBA1C has the mean normalized difference of 4.7% which was caused by the difference in the rounding. TSH and PHOS have known issues in the source data. Magnesium no longer appears in this figure.
Next, Figure 4 shows the normalized maximum differences between the source and the UCUM conversions when they are not equal. Now it becomes more clear that the maximum difference of BILI and MONOLE are relatively high. Both are LOINC based molar conversion and these were further invstigated.
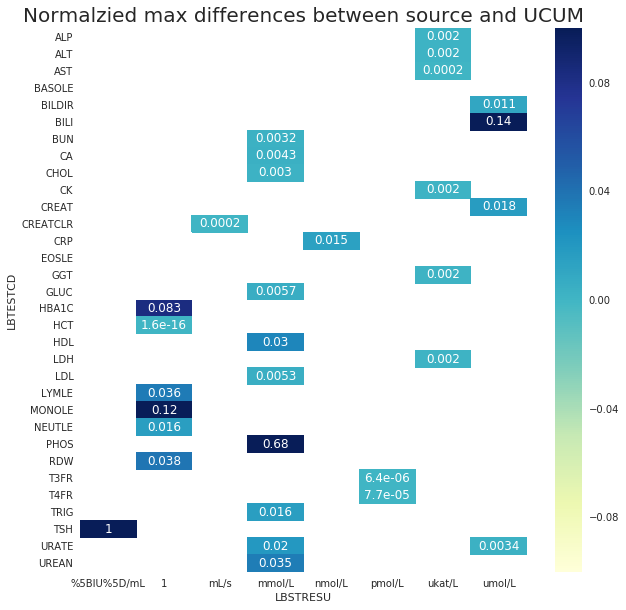
It was found that the differences in MONOLE were due to the rounding differences. On the other hand, the difference in BILI was not related to the rounding difference. For example, $0.4\frac{mg}{dL}$ was reported as 6.0 $\frac{\mu\text{mol}}{L}$
Per LOINC 1975-2, m.w is 584.6621g/mol for BILI, so $0.4\frac{mg}{dL}=0.04\frac{g}{L}*\frac{580*10e6 g}{\mu\text{mol}}=6.841558568615957\frac{\mu\text{mol}}{L}$. Therefore, the source data had incorrect conversions while UCUM conversions were correct.
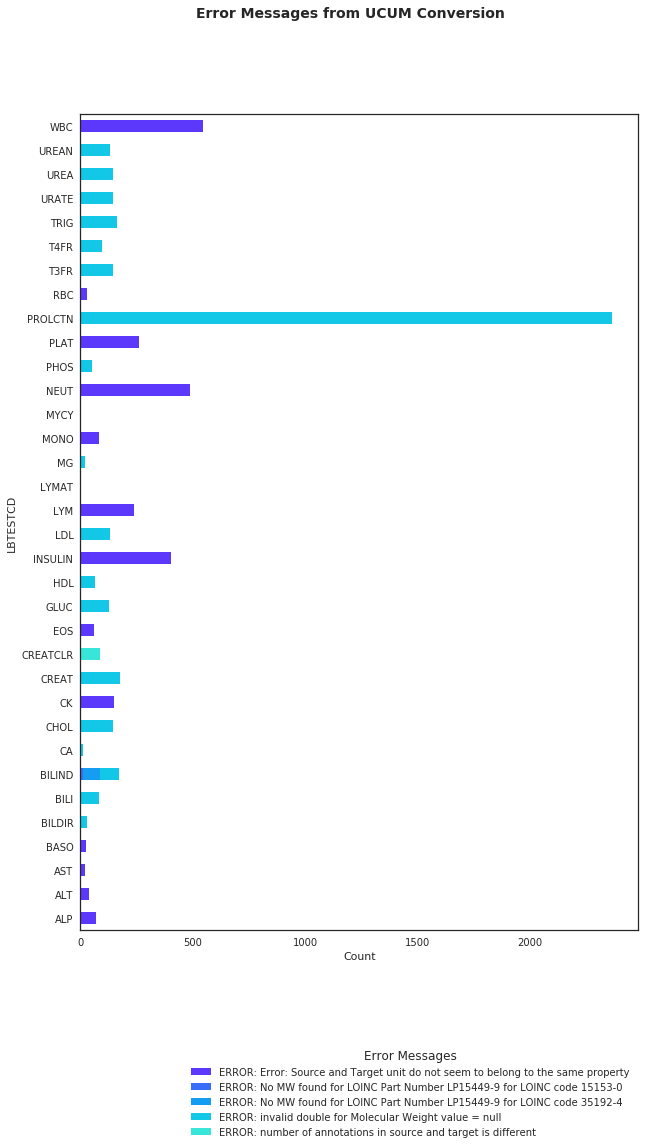
2-3-2. Error Messages from UCUM Conversion ¶
Figure 5 displays the distribution of error messages for each test.
- The most frequent error was
ERROR: invalid double for Molecular Weight value = nulldue to missing m.w. or LOINC when the conversion requires m.w.. - There were 2 kinds of
No MW found for LOINC Part Number LPxxxx for LOINC code yyyy. - Creatinine Clearance (mL/min/1.73 m2) conversion failed with
ERROR: number of annotations in source and target is different. The surface area 1.73 m2 was added as annotation for the source but the target did not include the same annotation. This will be solve with http://xml4pharmaserver.com:8080/UCUMService2/rest/ucumtransform/121/from/mL/min/%7B1.73_m2%7D/to/mL/s/%7B1.73_m2%7D - Several conversions failed with
ERROR: unexpected result: Error: Source and Target unit do not seem to belong to the same property. See Table 7 - 9 for more detail.
The figure below shows the error messages received from this exercise.
| response | note | checklist |
|---|---|---|
| ERROR: Error: Source and Target unit do not seem to belong to the same property | See details in Table 8 and 9 | http://xml4pharmaserver.com:8080/UCUMService2/rest/ucumtransform/12.9/from/uU/mL/to/pmol/L |
| ERROR: invalid double for Molecular Weight value = null | True: Due to missing MW in the source data | http://xml4pharmaserver.com:8080/UCUMService2/rest/ucumtransform/0.3/from/mg/dL/to/umol/L |
| ERROR: No MW found for LOINC Part Number LP15449-9 for LOINC code 15153-0 | True: Due to missing MW for LOINC | http://xml4pharmaserver.com:8080/UCUMService2/rest/ucumtransform/0.3/from/mg/dL/to/umol/L/LOINC/15153-0 |
| ERROR: No MW found for LOINC Part Number LP15449-9 for LOINC code 35192-4 | True: Due to missing MW for LOINC | http://xml4pharmaserver.com:8080/UCUMService2/rest/ucumtransform/1.07/from/mg/dL/to/umol/L/LOINC/35192-4 |
| ERROR: number of annotations in source and target is different | True: Due to not having the same annotations in the target | http://xml4pharmaserver.com:8080/UCUMService2/rest/ucumtransform/121/from/mL/min/%7B1.73_m2%7D/to/mL/s |
UCUM seems to have issue with G (giga) and T (Tera) even after the second letter was added (GA, TR). The ucum-essence.xml seems to suggest that %5BIU%5D i.e. [IU] and U are equivalent but the conversion was not successful.
| Index | LBTESTCD | LBORRESU | LBSTRESU |
|---|---|---|---|
| 5652 | INSULIN | uU/mL | pmol/L |
| 6586 | BASO | G/L | 10*9/L |
| 6588 | EOS | G/L | 10*9/L |
| 6591 | LYM | G/L | 10*9/L |
| 6594 | MONO | G/L | 10*9/L |
| 6597 | NEUT | G/L | 10*9/L |
| 6600 | PLAT | G/L | 10*9/L |
| 6606 | RBC | T/L | 10*12/L |
| 6609 | WBC | G/L | 10*9/L |
| 7026 | LYMAT | G/L | 10*9/L |
| 7828 | MYCY | G/L | 10*9/L |
| 11395 | ALP | %5BIU%5D/L | U/L |
| 11400 | ALT | %5BIU%5D/L | U/L |
| 11403 | AST | %5BIU%5D/L | U/L |
| 11416 | CK | %5BIU%5D/L | U/L |
| 15398 | INSULIN | u%5BIU%5D/mL | pmol/L</pre> |
| Index | LBTESTCD | LBORRESU | LBSTRESU |
|---|---|---|---|
| 5652 | INSULIN | uU/mL | pmol/L |
| 6586 | BASO | GA/L | 10*9/L |
| 6588 | EOS | GA/L | 10*9/L |
| 6591 | LYM | GA/L | 10*9/L |
| 6594 | MONO | GA/L | 10*9/L |
| 6597 | NEUT | GA/L | 10*9/L |
| 6600 | PLAT | GA/L | 10*9/L |
| 6606 | RBC | TR/L | 10*12/L |
| 6609 | WBC | GA/L | 10*9/L |
| 7026 | LYMAT | GA/L | 10*9/L |
| 7828 | MYCY | GA/L | 10*9/L |
| 11395 | ALP | %5BIU%5D/L | U/L |
| 11400 | ALT | %5BIU%5D/L | U/L |
| 11403 | AST | %5BIU%5D/L | U/L |
| 11416 | CK | %5BIU%5D/L | U/L |
| 15398 | INSULIN | u%5BIU%5D/mL | pmol/L</pre> |
2-3-3. Summary ¶
- many m.w. based conversions were successful when LOINC is provided
- the service can identify incorrect conversions provided by the vendors
- there are some challenges with units containing [IU], U, G, and T
- the conversion which involves 'Eq' was successful
[1] Wu and Wales (2017) Laboratory Data Standardization with SAS . PharmaSUG
[2] FDA (2013). Position on Use of SI Units for Lab Tests - FDA
[3] PMDA. (2015) Notification on Practical Operations of Electronic Study Data Submissions
[4] PMDA (2017). FAQs on Electronic Study Data Submission (Excerpt)
[5] Jozef Aerts (2019) SDTM --STRESN: why we need UCUM
Something to note:
The units used in API call has to be compliant with the USUM specifications. In addition, URL encoding has to be applied for some special characters. URL encoding can be found here.
#!pip install tqdm
import boto3
import botocore
import re
import os
import pandas as pd
#import pandas_profiling as pp
#import pixiedust as px
import pickle
import zipfile
from sas7bdat import SAS7BDAT
# Visual
import matplotlib.pyplot as plt
import seaborn as sns; sns.set()
%matplotlib inline
# my utilities
from lib.ucum import *
bucket='snvn-sagemaker-1' #data bucket
s3 = boto3.resource('s3')
url='http://xml4pharmaserver.com:8080/UCUMService2/rest'
new=0 # set to 1 if use new dataset
3-2. Copy data and unzip ¶
%%time
KEY=os.path.join('mldata','Sam','data','project','pool','lb.zip')
os.makedirs('data', exist_ok=True)
try:
s3.Bucket(bucket).download_file(KEY, os.path.join('data','lb.zip'))
except botocore.exceptions.ClientError as e:
if e.response['Error']['Code'] == "404":
print("The object does not exist.")
else:
raise
zip_ref = zipfile.ZipFile(os.path.join('data','lb.zip'), 'r')
zip_ref.extractall()
zip_ref.close()
3-3. Retain records for verification ¶
if new==1:
rlb=SAS7BDAT(os.path.join('data','lb.sas7bdat')).to_data_frame()
elif new!=1:
rlb=SAS7BDAT(os.path.join('data','raw_lb.sas7bdat')).to_data_frame()
print('Number of studies: ',len(list(set(rlb['STUDYID']))))
df=rlb[['LBTESTCD','LBTEST','LBORRES','LBORRESU','LBSTRESU','LBSTRESN','LBLOINC']]
#df=df[df['LBORRESU']!='LBSTRESU'] #since we don't need to verify
print('Number of records in the input: ',df.shape)
#Cleaning Input Data
df1=df.copy()
df1.dropna(axis=0, subset=['LBORRESU'], inplace=True)
df1.dropna(axis=0, subset=['LBSTRESU'], inplace=True)
print('Removed records with missing units: ', df1.shape)
#Remove records not needed for verification
#1. results contain character
df1['LBORRES']=df1['LBORRES'].str.replace(r'[a-zA-Z ]+','')
df1=df1[df1['LBORRES']!='']
df1.dropna(axis=0, subset=['LBSTRESN'], inplace=True)
print('Removed records with character results: ',df1.shape)
#2. both units are the same
df1=df1[df1['LBORRESU']!=df1['LBSTRESU']]
print('Removed records do not require conversion: ',df1.shape)
df1.to_csv('lb.csv')
Let's see what units have been used in the input datasets
bar_hm(df1,'Units found in the input data (counts)')
3-4. Define regular expression used to convert the input units to UCUM ¶
#Regular expressions. --- update this based on raw data
patterns = [("%","%25"),
("\A[xX]?10[^E]", "10*"),
("IU", "%5BIU%5D"),
("\Anan", ""),
("\ANONE", ""),
("\A[rR][Aa][Tt][Ii][Oo]", ""),
("\ApH", ""),
("Eq[l]?","eq"),
("\ATI/L","TR/L"),#update T to TR
("\AGI/L","GA/L"),#update G to GA
("V/V","L/L"),
("[a-z]{0,4}/HPF","/%5BHPF%5D"),
("[a-z]{0,4}/LPF","/%5BLPF%5D"),
("fraction of 1","1"),
("sec","s"),
("1.73m2","%7B1.73_m2%7D"),
("\AG/L","GA/L")
]
3-5. Functions Developed ¶
- cleanlist: a helper function which takes the elements of patterns to output UCUM conformant unit
- orresu2ucum: a function which takes the dataframe containing LBORRESU and LBSTRESU, and apply
cleanlist - ucumVerify: a function which takes the list of units to verify the conformance to UCUM using
isValidUCUMand returns a list with either True or False. - convert_unit: a function which takes the dataframe containing LBORRESU and LBSTRESU in UCUM and returns a dataframe where original LBSTRESN is not equal to LBSTRESN from the UCUM-LHC Converter
3-6 Verify converted UCUM ¶
dfconverted, ucumlist=orresu2ucum(df1,patterns)
ucumVerify(ucumlist, url)
bar_hm(dfconverted,'UCUM found in the input data (counts)')
3-7. Run UCUM Conversion ¶
nodupdf=df1.drop_duplicates(subset=['LBTESTCD','LBORRESU','LBORRES','LBSTRESN','LBSTRESU'], keep='first')
print('Removed duplicate records: ', nodupdf.shape[0])
nodupdf.shape
if os.path.isfile('output/findingsv2.pickle'):
findings= open('output/findingsv2.pickle', mode='rb')
findings=pickle.load(findings)
full= open('output/fullv2.pickle', mode='rb')
full=pickle.load(full)
response= open('output/responsev2.pickle', mode='rb')
response=pickle.load(response)
else:
findings,full,response=convert_unit(nodupdf, url, patterns,loinconly=0)
with open('output/findingsv2.pickle', 'wb') as handle:
pickle.dump(findings, handle, protocol=pickle.HIGHEST_PROTOCOL)
with open('output/fullv2.pickle', 'wb') as handle:
pickle.dump(full, handle, protocol=pickle.HIGHEST_PROTOCOL)
with open('output/responsev2.pickle', 'wb') as handle:
pickle.dump(response, handle, protocol=pickle.HIGHEST_PROTOCOL)
4. Output Issues ¶
4-1. Discrepancies between LBSTRESN and Conversion based on UCUM ¶
findings[(findings['fromucum'].notnull())]
discrepant=findings[(findings['fromucum'].notnull())]
diff=discrepant.copy()
diff['diff']=np.array(discrepant['LBSTRESN'])-np.array(discrepant['fromucum'])
diff['diffpct']=abs(np.array(discrepant['LBSTRESN'])-np.array(discrepant['fromucum']))/np.array(discrepant['LBSTRESN'])
miss=diff[diff['diff']==np.nan]
diff.dropna(axis=0, how='any',subset=['diff'], inplace=True)
dfpiv=diff.pivot(columns='LBTESTCD', values='diff')
difftests=len(dfpiv.columns)
review=diff[abs(diff['diffpct'])>0.01]
diff.to_csv('diff.csv')
miss.to_csv('miss.csv')
discrepant0=diff.groupby(by=['LBTESTCD','LBSTRESU'],as_index =True)
discrepant0=discrepant0.describe().xs('mean', level=1, axis=1)[['LBSTRESN','diffpct']]
discrepant0.reset_index(inplace=True)
sns.set(style='white')
plt.subplots(figsize=(10,10))
ax = sns.heatmap(discrepant0.pivot('LBTESTCD','LBSTRESU','diffpct'),annot=True, cmap="YlGnBu")
ax.axes.set_title('Normalzied mean differences between source and UCUM',fontsize=20)
Note that MG is disappeared
4-2-2. Normalized maximum differences between souce and UCUM ¶
discrepant0=diff.groupby(by=['LBTESTCD','LBSTRESU'],as_index =True)
discrepant0=discrepant0.describe().xs('max', level=1, axis=1)[['LBSTRESN','diffpct']]
discrepant0.reset_index(inplace=True)
import matplotlib.pyplot as plt
plt.subplots(figsize=(10,10))
ax2 = sns.heatmap(discrepant0.pivot('LBTESTCD','LBSTRESU','diffpct'),annot=True, cmap="YlGnBu")
ax2.axes.set_title('Normalzied max differences between source and UCUM',fontsize=20)
Normalized max difference of >10% observed in BILI, MONOLE, MG, PHOS, and TSH. PHOS, and TSH have known issues.
4-2-3. Normalized minimum differences between souce and UCUM ¶
discrepant0=diff.groupby(by=['LBTESTCD','LBSTRESU'],as_index =True)
discrepant0=discrepant0.describe().xs('min', level=1, axis=1)[['LBSTRESN','diffpct']]
discrepant0.reset_index(inplace=True)
plt.subplots(figsize=(10,10))
ax3 = sns.heatmap(discrepant0.pivot('LBTESTCD','LBSTRESU','diffpct'),annot=True, cmap="YlGnBu")
ax3.axes.set_title('Normalzied min differences between source and UCUM',fontsize=20)
discrepant0=diff.groupby(by=['LBTESTCD','LBSTRESU'],as_index =True)
discrepant0=discrepant0.describe().xs('mean', level=1, axis=1)[['LBSTRESN','diff']]
discrepant1=pd.melt(discrepant0.reset_index(), id_vars=['LBTESTCD','LBSTRESU'],value_vars=['LBSTRESN','diff'])
4-2-4. Compare mean results vs. mean differences ¶
Left bar = mean, right bar = mean diff
g = sns.FacetGrid(discrepant1, col='LBSTRESU', row='LBTESTCD', sharey='row', margin_titles=True) g.map(sns.barplot, 'LBTESTCD', 'value', 'variable', hue_order=['LBSTRESN','diff'], alpha=.8)
issues=['BILI','HBA1C','MONOLE','MG','PHOS','TSH']
discrepant2=discrepant1[discrepant1['LBTESTCD'].isin(issues)]
g = sns.FacetGrid(discrepant2, col='LBSTRESU', row='LBTESTCD', sharey='row', margin_titles=True)
g.map(sns.barplot, 'LBTESTCD', 'value', 'variable', hue_order=['LBSTRESN','diff'], alpha=.8)
g.fig.suptitle('mean (left) and mean differences (right) between source and UCUM',y=1.02)
Note: Issues were identified in MG, PHOS, and TSH. Other differences are neglibile.
#pp.ProfileReport(dfpiv)
#px.display(dfpiv)
4-2-5. Check distributions of differences between the source and UCUM conversion ¶
import random
c_=[]
for i in range(27):
r = lambda: random.randint(0,255)
c='#%02X%02X%02X' % (r(),r(),r())
c_.append(c)
fig = plt.figure( figsize=(20,20))
fig.subplots_adjust(hspace=0.4, wspace=0.4)
idx = np.arange(1, difftests+1)
for i, col, c in zip(idx, dfpiv.columns, c_):
ax = fig.add_subplot(7, 7, i)
#dfpiv.loc[:, col].plot.hist(label=col, color=c, range=(diff['diff'].min(), diff['diff'].max()), bins=15)
dfpiv.loc[:, col].plot.hist(label=col, color=c, bins=20)
plt.yticks(np.arange(0, 100, 10))
plt.suptitle('Distributions of differences between LBSTRESN and UCUM conversion. \nExcluding exact match between LBSTRESN and UCUM conversion', fontsize=14, fontweight='bold')
plt.legend()
4-2-6. Summary statistics of the source and UCUM conversion on $\text{Mg}^{+2}$ ¶
There was no difference between the souce and UCUM, so this step is skipped.
#check=sumstat('MG',findings, rlb, dfpiv)
#check.head()
4-2-7. Summary statistics of the source and UCUM conversion on PHOS ¶
check=sumstat('PHOS', findings,rlb, dfpiv)
check.head()
Phosphate molecular weight is 94.97g/mol 3.0mg/dL in mmol/L is
$\frac{3.0\text{mg}}{\text{dL}} =\frac{30\text{mg}}{\text{L}} = \frac{0.03\text{g}}{\text{L}}*\frac{\text{mmol}}{0.09497\text{g}} = 0.3158892281773191 $
(3*10/1000)/(94.97/1000)
Note: The identified error is originated from the lab vendor.
4-2-8. Summary statistics of the source and UCUM conversion on TSH ¶
check=sumstat('TSH',findings, rlb, dfpiv)
check.head()
m[IU]/L to [IU]/mL
$\frac{\text{mIU}}{\text{L}}=\frac{\text{IU}}{1000\text{L}}=\frac{\text{IU}}{10^6\text{mL}}$
1.01/10**6
Note: The identified error is originated from the lab vendor.
4-2-9. Summary statistics of the source and UCUM conversion on BILI ¶
check=sumstat('BILI',findings, rlb, dfpiv)
check.head()
Per LOINC m.w is 584.6621g/mol
so $0.4\frac{mg}{dL}=0.04\frac{g}{L}*\frac{580*10e6}{\mu\text{mol}}=6.841558568615957$
UCUM is correct.
0.04/(584.6621*10e-6)
4-2-10. Summary statistics of the source and UCUM conversion on MONOLE ¶
check=sumstat('MONOLE',findings, rlb, dfpiv)
max diff is found in the last record...
mo_=diff[diff['LBTESTCD']=='MONOLE']
mo_[mo_['diffpct']==mo_['diffpct'].max()]
4-2-11. Summary statistics of the source and UCUM conversion on HBA1C ¶
check=sumstat('HBA1C',findings, rlb, dfpiv)
mo_=diff[diff['LBTESTCD']=='HBA1C']
mo_[mo_['diffpct']==mo_['diffpct'].max()]
Looks like the difference is based on rounding
4-3. List of records in which UCUM conversion were not sucessful ¶
ucumfail=findings[(findings['fromucum'].isnull())]
ucumfail
errormsg=ucumfail.dropna(subset=['response'])
errormsg=errormsg.groupby(['LBTESTCD','response']).count().iloc[:,0].reset_index()
errormsg.columns=['LBTESTCD','RESPONSE','COUNT']
fig1 = plt.figure( figsize=(10,10))
sns.set_palette("rainbow", 13)
ax4 = errormsg.pivot('LBTESTCD','RESPONSE','COUNT').plot(kind='barh',figsize=(10,15),stacked=True)
plt.legend(title='Error Messages',loc=5, bbox_to_anchor=(1, -0.2))
plt.suptitle('Error Messages from UCUM Conversion', fontsize=14, fontweight='bold')
plt.xlabel("Count")
ucumfail.dropna(subset=['response']).to_csv('errors.csv')
4-3-1. Summary of Error Message finding ¶
msglist=list(set(ucumfail['response']))
for i in range(len(msglist)):
e_=ucumfail['response']==msglist[i]
o_=ucumfail[e_][['LBTESTCD','LBORRESU','LBSTRESU','checklist','response']].drop_duplicates(subset=['LBTESTCD','LBORRESU','LBSTRESU'])
print('Error: '+msglist[i])
print('Call: '+o_['checklist'])
print()
print(o_[['LBTESTCD','LBORRESU','LBSTRESU']])
print()
print('---------------------------------------------------------------------------------------')
pd.options.display.max_colwidth =150